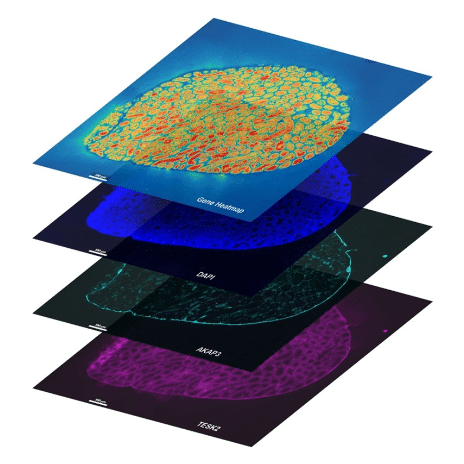
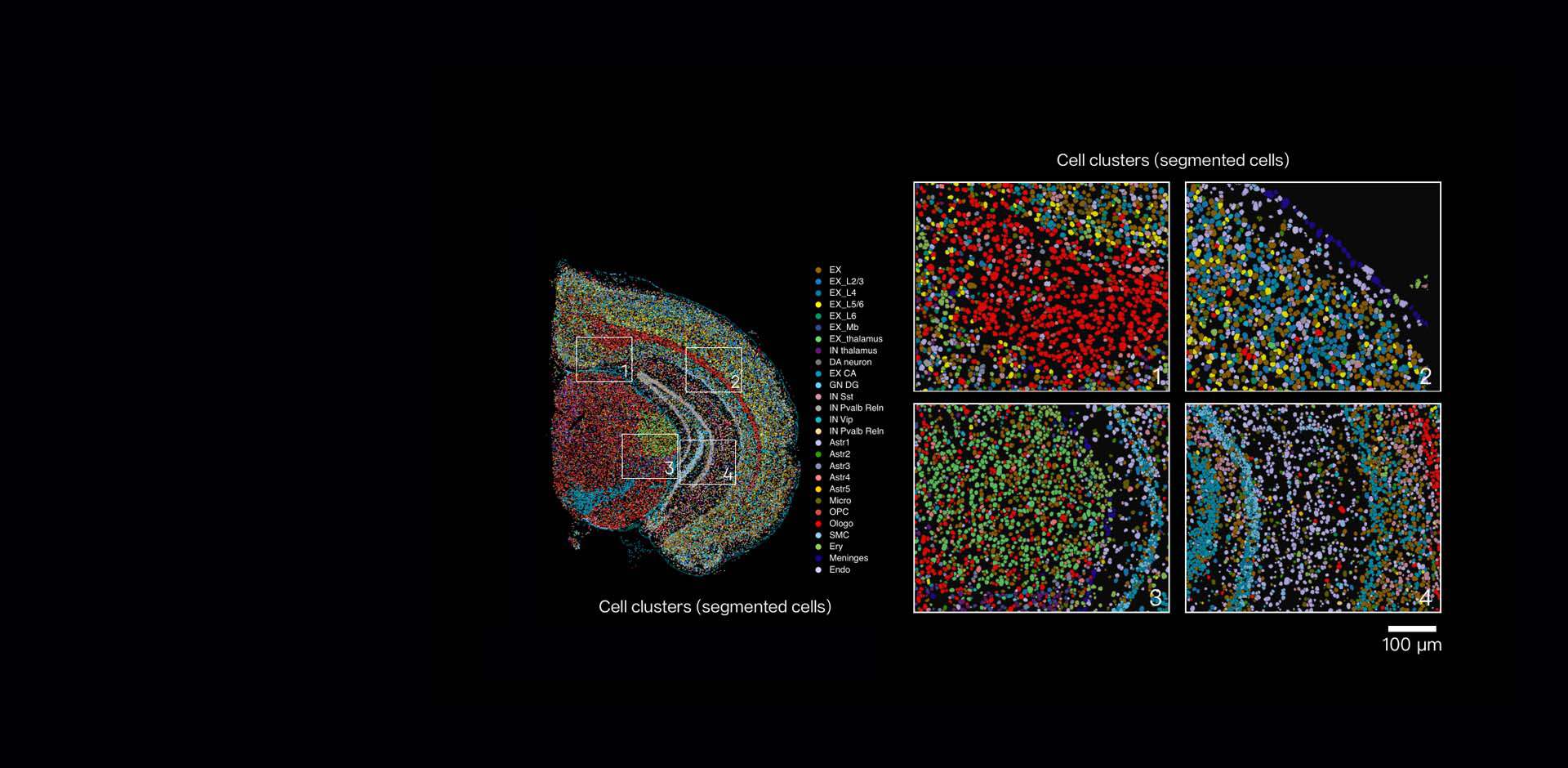
Spatial transcriptomics was named the method of the year in 2020 (Nature 2021). Spatial transcriptomics has rapidly emerged as a transformative technology in molecular biology, providing unprecedented insights into gene expression within the spatial context of tissues. From advancing fundamental biology to enabling precision medicine, spatial transcriptomics redefines how scientists approach cellular biology. Key technologies like Stereo-seq, 10x Genomics Xenium, and Akoya Biosciences platforms are at the forefront of this innovation, each bringing unique capabilities to the field.
Unlocking Spatial Genomics with Stereo-seq
Stereo-seq (SpaTial Enhanced REsolution Omics-sequencing), developed by BGI-Research, offers a sequencing-based approach to spatial transcriptomics. Unlike probe-based systems, Stereo-seq captures any protein-coding or lncRNA transcript from any organism, providing unparalleled versatility. This flexibility has positioned Stereo-seq as a powerful tool for diverse research applications, from plant biology to human disease studies.
MiRXES, a pioneering provider of next-generation sequencing services, was among the first certified stereo-seq service providers and remains one of only two providers in the United States. Researchers leveraging Stereo-seq benefit from its capacity to map complex transcriptomes at subcellular resolution, delivering detailed insights into tissue architecture and cellular interactions.
High-Plex Detection with 10x Genomics Xenium
10x Genomics has also made significant contributions with its Xenium platform, which includes the Xenium Prime 5K pan-tissue kit. Xenium’s high-plex capabilities have enabled researchers to explore intricate tissue architectures and identify gene expression patterns tied to health and disease. For applications where probe specificity aligns with research goals, Xenium remains a powerful platform for spatial transcriptomics.
Xenium can detect up to 5,000 genes and offers high-plex gene expression profiling with robust performance for human and mouse samples. However, its reliance on probe-based detection limits its use to other model organisms and for studies requiring analysis of the entire transcriptome.
Multiplex Imaging with Akoya Biosciences
Akoya Biosciences specializes in spatial phenotyping through high-parameter tissue imaging. Its platforms integrate spatial biology and multiplexed imaging, enabling visualization of protein expression alongside RNA. By complementing sequencing-based approaches like Stereo-seq and Xenium, Akoya’s solutions provide a holistic view of tissue biology, helping researchers link molecular mechanisms to phenotypic outcomes. However, Akoya’s technology depends on phenocode signature panels, limited to only a few dozen genes and not yet wholly optimized for non-human samples.
Spatial Transcriptomics and the Human Cell Atlas
The Human Cell Atlas (HCA) initiative aims to map every cell type in the human body, creating a comprehensive reference for human biology and disease. Spatial transcriptomics plays a vital role in this endeavor by adding spatial context to cellular atlases, revealing how cells interact within tissues. Another technology the HCA consortium uses is single-cell whole genome sequencing (scWGS). One of the significant obstacles in the past for scWGS has been the challenge of getting complete genome coverage from a single cell. This has largely been solved with the advent of Primary Template-directed Amplification (PTA), which produces significantly improved and reproducible genome sequencing coverage and variant detection from a single genome of a single cell. MiRXES was one of the first service providers to use PTA for scWGS.
Spatial Transcriptomics: Transforming Health and Disease Research
Spatial transcriptomics is redefining the understanding of health and disease by mapping gene expression within the intricate spatial architecture of tissues. This cutting-edge technology allows scientists to unravel how cells communicate and function in their native environments, advancing insights into complex diseases such as cancer, autoimmune disorders, and kidney disease.
Recent studies demonstrate the profound impact of spatial transcriptomics in medicine. For example, researchers have used this technology to identify cellular interactions driving kidney fibrosis, providing potential therapeutic targets for chronic kidney disease (Nature Reviews Nephrology, 2024). Spatial transcriptomics has also been used to uncover tumor aging microenvironment patterns in prostate cancer, analyze diabetic kidney biopsy gene expression, and reveal spatial heterogeneity in central nervous system lymphomas. These findings demonstrate its potential to inform personalized diagnostics and treatment strategies by linking molecular and spatial data. (Theranostics, 2024).
Moreover, applications in neuroscience uncover gene expression patterns that underlie neurological disorders. A notable study published last week (Nature Communications 2025) used Stereo-seq to construct a subcellular-resolution spatial transcriptome atlas of the human prefrontal cortex (PFC) from Alzheimer’s disease patients. The study identified critical gene expression modules within the PFC and revealed ZNF460 as a transcription factor regulating these modules. This discovery highlights a potential therapeutic target for neurological disorders, underscoring the transformative potential of Stereo-seq in advancing human health.
As spatial transcriptomics continues to evolve, its integration with other multi-omics approaches promises to illuminate the molecular landscapes of health and disease, paving the way for precision medicine. By bridging cellular biology and spatial context, this revolutionary field is unlocking new therapeutic possibilities across a spectrum of diseases.
Future Directions and Challenges
The rapid development of spatial transcriptomics technologies has created a vibrant ecosystem of tools and applications. However, challenges remain, including standardizing data analysis pipelines, integrating multi-omics datasets, and ensuring cost accessibility for broader adoption.
Innovations like Stereo-seq’s sequencing-based approach and Akoya’s multiplex imaging address these challenges by providing scalable and versatile solutions. As platforms like 10x Genomics Xenium continue to enhance their capabilities, the field is poised to deliver even deeper insights into cellular and tissue biology.
Spatial transcriptomics is revolutionizing our understanding of biology by bridging molecular data with spatial context. From enabling the Human Cell Atlas to uncovering therapeutic targets in the brain, technologies like Stereo-seq, 10x Genomics Xenium, and Akoya Biosciences are shaping the future of biomedical research. As advancements continue, spatial transcriptomics promises to unlock new frontiers in health and disease, making it one of the most exciting areas in life sciences today.
Owen Howard, an undergraduate at Florida State University, FSU Department of Biology, wrote this article.